 |
PixCells
This project is a Cellular Potts Model (CPM) simulator. It was developped during a Master's degree in Physics as a master project.
|
 |
PixCells
This project is a Cellular Potts Model (CPM) simulator. It was developped during a Master's degree in Physics as a master project.
|
This project is a simulation of cells on a lattice using the Cellular Potts Model (CPM). The CPM is a lattice-based model that allows the simulation of cells and their interactions. The cells are represented by a set of connected pixels on a lattice. The model is based on the Metropolis algorithm, where the energy of the system is minimized by moving the cells on the lattice. The energy of the system is given by a Hamiltonian that takes into account the interactions between the cells and the properties of the cells. The model can be used to study the behavior of cells in different environments and to investigate the mechanisms of cell migration, sorting, and aggregation.
Moreover, this project has the implicit goal to simulate vascularisation of tissues. The cells are moving on a lattice and are interacting with each other. They can elongate, contract, and move. The cells are also interacting with the extracellular matrix (ECM) and with a chemoattractant molecule. The cells are also connected to each other by a force that tries to keep them together.
For more details on the theory behind the model, you can check the Theory page.
The model is implemented in C++ and visualized using the SFML library. The cells are represented by colored pixels on a lattice, and different cell's id are visualized by a border. The simulation can be run with different parameters to study the behavior of cells in different environments.
Here we can see an exemple to get a vascular network. Everything can be changed in the config.txt file.
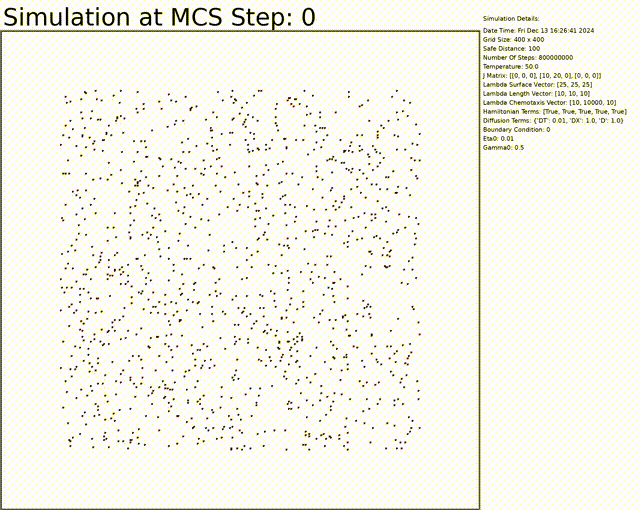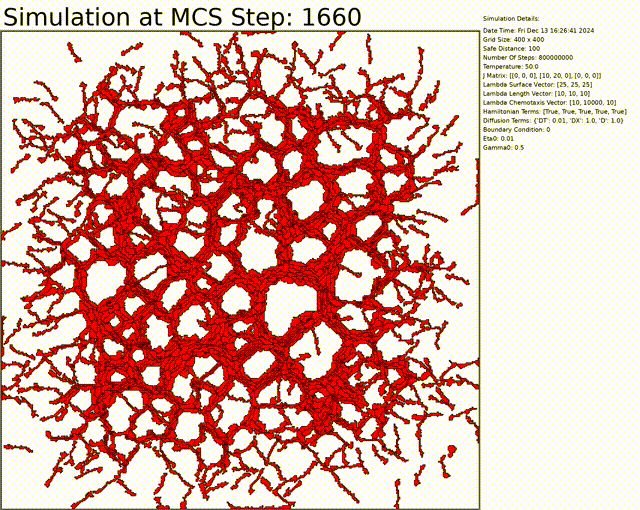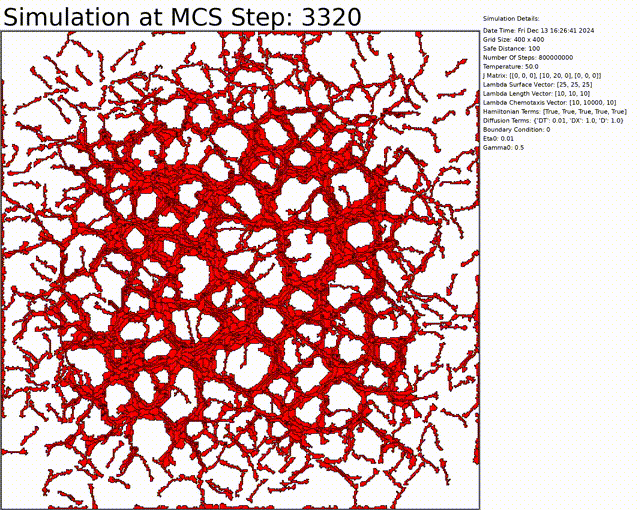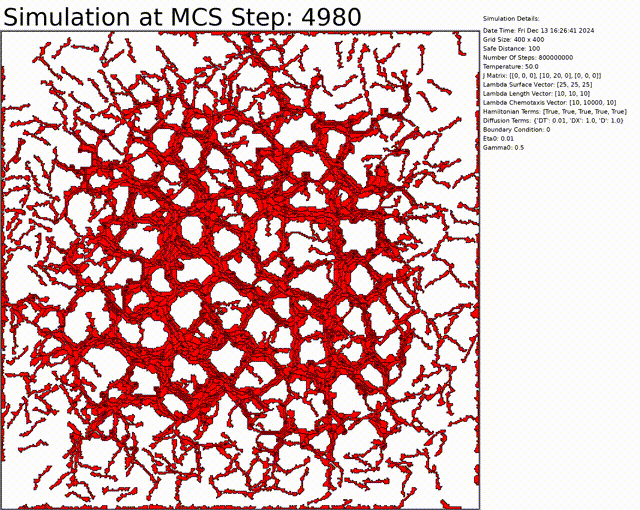
You can download the code by cloning the repository:
Then go in the directory:
You will need to create a data directory and some image and video directories:
You will need to install the dependencies (if not installed yet):
If not already made, you should use a virtual environment for the python script:
Then you need to install all the libraries needed for the python script:
Then you can compile the code with the following command:
To create the Doxygen documentation, you can run the following command:
To create a tar.gz file with the code and the documentation, you can run the following command:
You should check the Configuration file page to see how to create a config.txt file.
You can run the code with:
or
for the default config file config.txt and output file data.txt with a render activated.
You can also run the code yourslef with the following command:
And finally you can run the python script:
Then, you can create a video with the following command:
or a gif with ffmpeg:
The ./run_config_and_plot.sh script will do all of this for you.
You can contact me at lucas.nosp@m.bois.nosp@m.tay@p.nosp@m.roto.nosp@m.n.me for any question or suggestion.
I'm eager to any contribution, feel free to make a pull request. The system is not perfect and can be improved in many ways. Moreover, a lot of features can be added to the simulation.
This work © 2024 by Lucas BOISTAY is licensed under Creative Commons Attribution-NonCommercial 4.0 International.