|
PixCells
This project is a Cellular Potts Model (CPM) simulator. It was developped during a Master's degree in Physics as a master project.
|
 |
PixCells
This project is a Cellular Potts Model (CPM) simulator. It was developped during a Master's degree in Physics as a master project.
|
The goal here is to simulate a sorting of cell. The interaction matrix \( J \) has lower values on the diagonal and higher values on the off-diagonal. This means that cells tends to be together if they are the same type and tend to be separated if they are different.
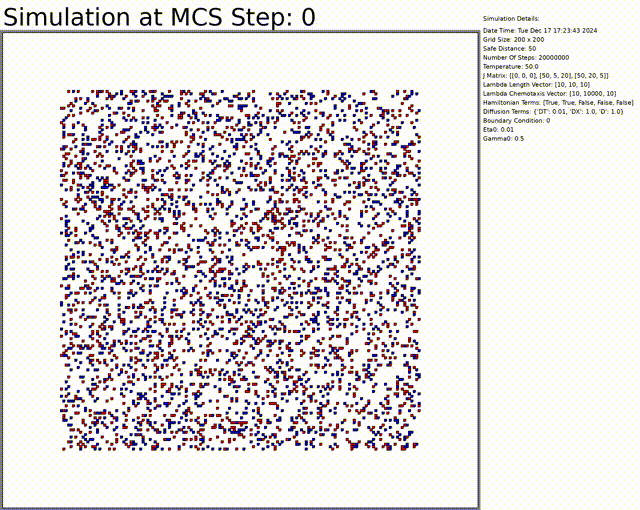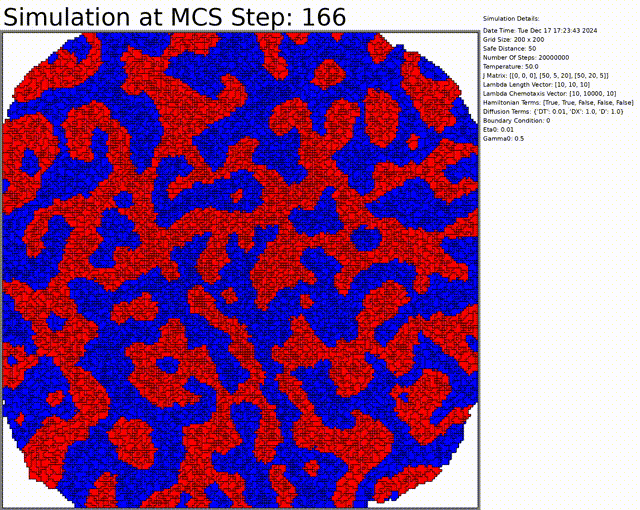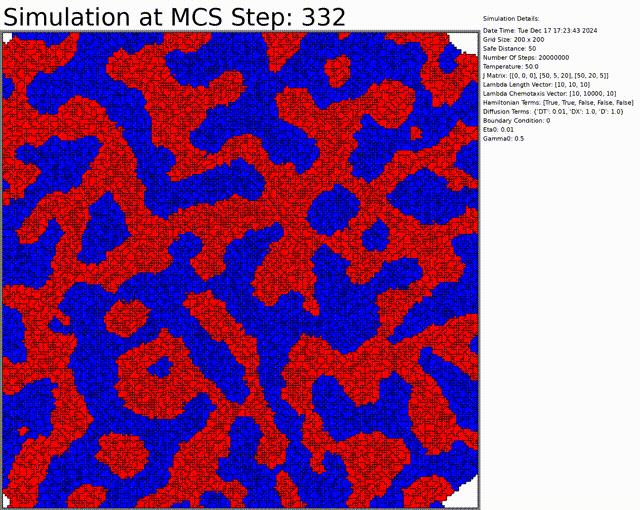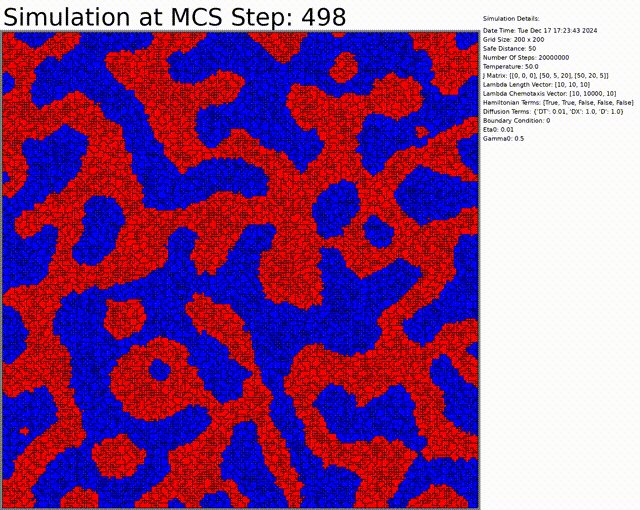
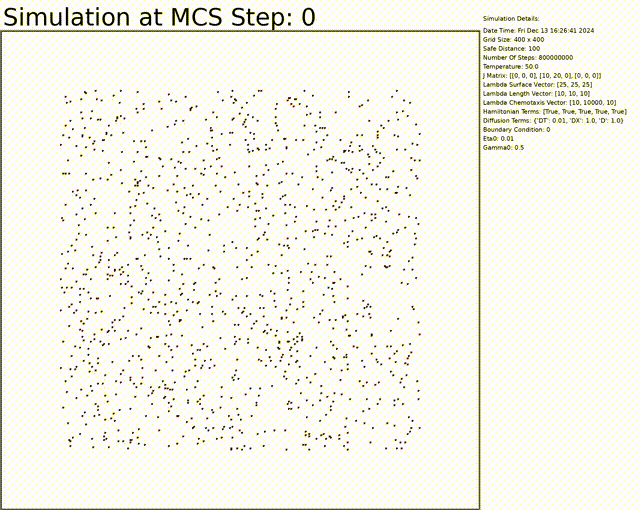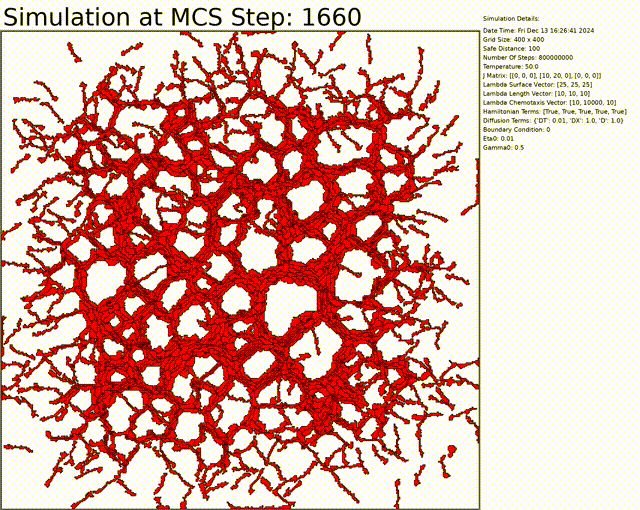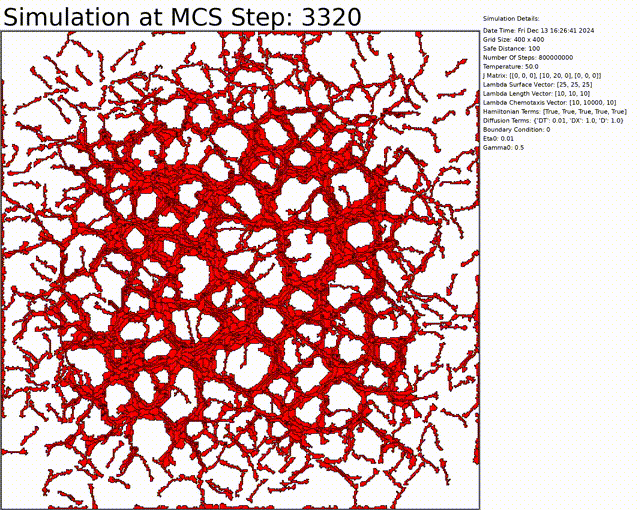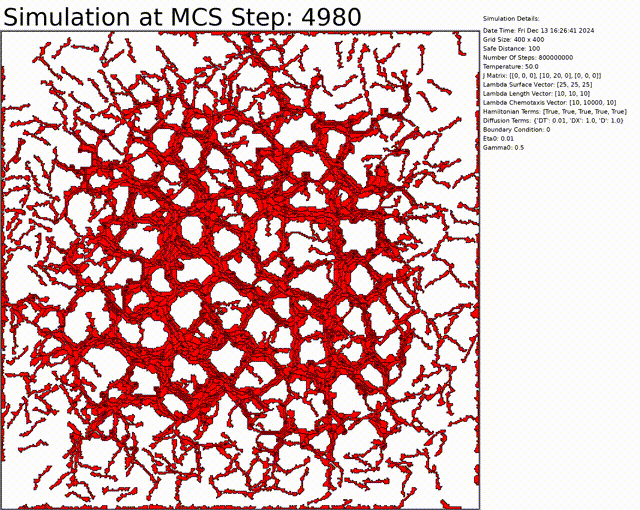
The goal here is to simulate a network of cells. The chemotaxis, length, and connectivity are activated. The cells balance between the chemotaxis that attracts them to one another, and the interaction that tends to separate them at low distances. This results in a beautiful network of cells.